Research Complex at Harwell provides support in identifying the replication cycle of COVID-19
Research conducted by Central Laser Facility, Diamond Light Source and the Universities of Pittsburgh, Oxford, and Cambridge has examined how the COVID-19 virus reproduces.
This study, published in Nature Communications, is the first to examine how the virus's replication cycle is structured using the serial CryoFIB/SEM volume imaging facilities at Research Complex.
Since the outbreak of the COVID-19 pandemic, there has been intense research on purified viral components and inactivated viruses to help assess how the COVID-19 virus is structured. However, evidence on how the infection progresses is scarce, and there has been a lack of comprehensive knowledge on how the virus replicates.
Researchers from CLF's OCTOPUS Imaging Cluster, based at Research Complex, were among the first at Harwell Campus to have rapid access to study the virus. Using the Zeiss Crossbeam 550XL at Research Complex, fitted with a Quorum transfer station and cryo-stage, cells infected with the COVID-19 virus were examined at -165 celsius. Previous investigations have examined Leigh Syndrome, where scientists from CLF sliced parts of the cell using a focus ion beam (FIB) and employed a scanning electron microscope (SEM) to take 2D images of the cells. These sections of the cell were then reconstructed to form 3D images. The 3D image of the infected cells demonstrated that COVID-19 has visible structural alterations, like extensive membrane tunnels, formed for the virus to spread with lesions near the exit region of the cell. In order to obtain an in-depth perspective of the activity within the macromolecular complexes of an infected cell, another imaging technique named CryoET was used.
'Combined together, these provide a complementary view of the virus's activity. CryoFIB/SEM and CryoET revealed that the virus replicates in a highly organised and efficient manner. These were the steps of the virus replication cycle:
- RNA Synthesis and Transport: The first step is replication of the viral RNA. The replication takes place in an enclosed space called a double membrane vesicle (DMV). Viral RNA (vRNA) is packaged into a DMV, so it evades a patient's immune system.
- Assembly and Budding: vRNA being produced pushes other structural proteins to be formed (ex. N,M,E and S proteins) at the protein production site. The virus spikes, which give the virus its signature shape are also produced at this site.
- Assembly and Budding: Structural proteins, spikes and vRNA fuse together and are enclosed in a space referred to as a single membrane vesicle. Through further production of the structural elements, single membrane vesicles containing the virus become large virus contained vesicles.' (CLF News, 30th July 2021).
This integrated approach allows a holistic view of the COVID-19 infection, from the whole cell to individual molecules and signifies the importance of collaborative work at Harwell to fight the virus.
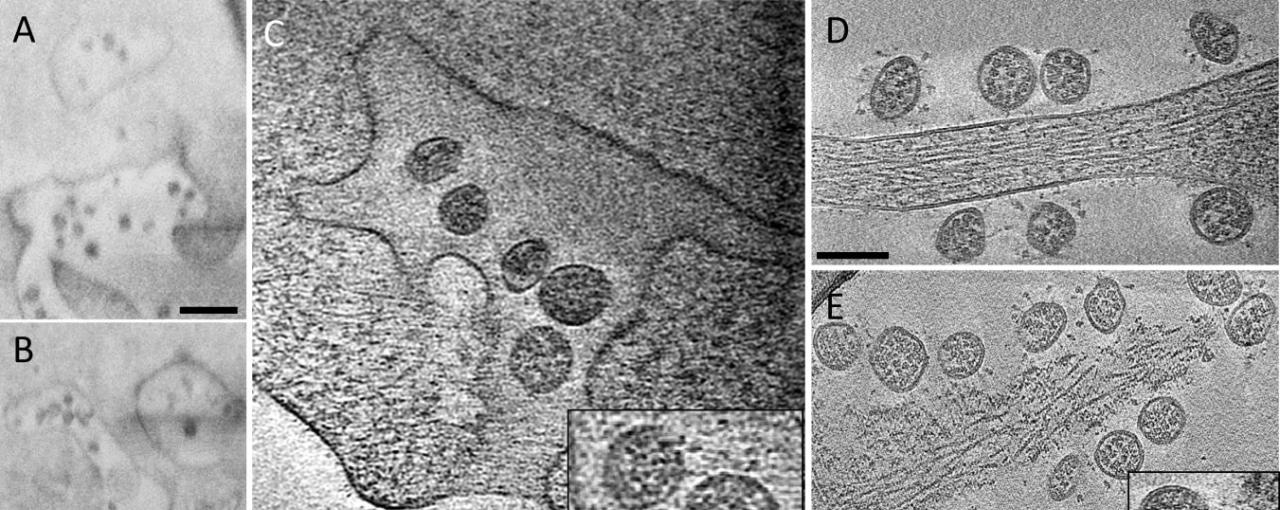
Image: Fig 5 - A, B CryoFIB/SEM images of cell periphery, depicting virus particles exiting through extended tunnels connected to external of the cell. C CryoET of the SARS-CoV-2-exiting tunnel. Inset shows a close-up view of viruses with prefusion spikes (red arrows). D, E Viruses released from the cell, close to an intact cell membrane (D) or next to membrane lesion sites (E). Inset, close-up view of a membrane lesion site. F Subtomogram average of spikes on released viruses at 10.6 Å resolution fitted with an atomic model of spike trimer (PDB 6VXX)16, viewed from side and top. G Comparison of spike structures from intracellular assembled viruses (transparent blue) and extracellular released viruses (transparent gray), viewed from top. Scale bar is 300 nm in A, B and 100 nm in C–E.