We are currently witnessing a rapid growth in the number of structures determined using cryogenic Electron microscopy (cryoEM). The Collaborative Computational Project for Electron Cryo-Microscopy, or CCP-EM, specializes in precisely this area of research. Based at Research Complex, they aim to provide software solutions to support the growing use of Cryo-EM in structural biology.
At intermediate-to-low resolutions, flexible fitting is typically adopted to place individual biomolecule structures into the cryoEM map. Identifying rigid bodies in biomolecules can guide this process of flexible fitting to help reach an optimum solution. Doing so can both speed up, and improve the accuracy of models.
While there already exist tools to identify rigid bodies in protein structures, these have previously been lacking for nucleic acid structures.
Alongside colleagues from the Leibniz Institute of Virology, and Birkbeck University of London, the CCP-EM group has developed a software tool, named RIBFIND2, in order to identify rigid bodies in nucleic acid. The first iteration of this tool, RIBFIND, clustered protein secondary structures into rigid bodies. In its second generation, researchers will now be able to use this tool to advance their models of RNA structures.
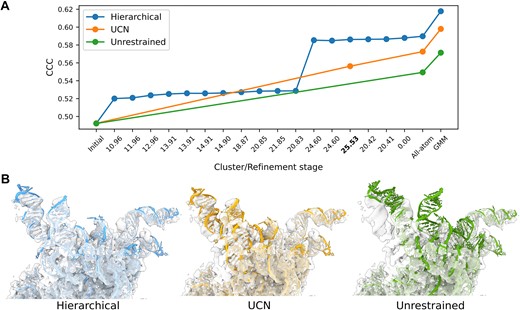
In a recent paper published in Nucleic Acids Research, the new software is tested across various examples containing RNA and protein. The team used the hierarchical approach, which leads to progressively smaller clusters and thus more and more flexibility. The fitting scores and RMSD (to the known ‘target’ structure) were best for the hierarchical approach, suggesting a clear advantage over unrestrained refinement.
Lead author Sony Malhotra says:
“We have expanded the original RIBFIND algorithm and now it can be applied to structures containing protein and/or RNA. The software is freely available both as a webserver and a standalone program. We have also implemented the tool in the CCP-EM software suite for wider dissemination.”
Co-author Agnel Joseph adds:
‘It is quite a powerful tool to tackle model building with low resolution cryoEM data. RIBFIND has wide range of applications: from identifying compact substructures to sampling biomolecular conformational changes.”
The full paper can be read here.



