Efforts led by scientists at the Research Complex at Harwell, Diamond Light Source and University of Oxford have revealed new interactions in the structure of Nontypeable Haemophilus influenzae (NTHi) SapA to help fight against respiratory illness, opening up the pathway for new research to battle the third leading cause of death globally.
Respiratory illness currently ranks as the third leading cause of death globally, with rates peaking for the very young and the elderly. Nontypeable Haemophilus influenzae (NTHi) is a significant respiratory tract pathogen which lives in the human upper respiratory tract and is common in many infections such as sinusitis, pneumonia and secondary chronic respiratory disease. In order for NTHi to continue colonizing within the host, it needs to survive by using up nutrients as the host’s immune system tries to respond. Outside of the respiratory context, NTHi is also a major pathogen in meningitis and otitis media, with the latter being a leading cause of specifically treated disease in children both in the UK and worldwide.
The high incidence of this bacterium exacerbating illnesses makes is a strategic target for biochemical investigation. While antibiotics can be effective in treating NTHi, the bacterium is nevertheless gaining resistance to other antibiotic classes. Ampicillin resistant strains of NTHi now exceed 30% in some countries, which has inevitably led to higher rates of treatment failure, increased costs, and decreased availability for severely or chronically ill patients.
An essential system for the survival of NTHi in its human host is the Sap (sensitivity to antimicrobial peptides) ABC transporter system. This system has been previously proposed to carry iron in the form of heme across the bacterial membrane. Once inside the cell, iron is used in bacterial nutrition. The same system has also been suggested to carry host antimicrobial peptides (AMPs). AMPs are part of the human immune system and work against invading bacteria by disrupting their membranes. By carrying the AMPs, the Sap system sequesters the damaging AMPs away from the membranes, keeping the bacterium protected.
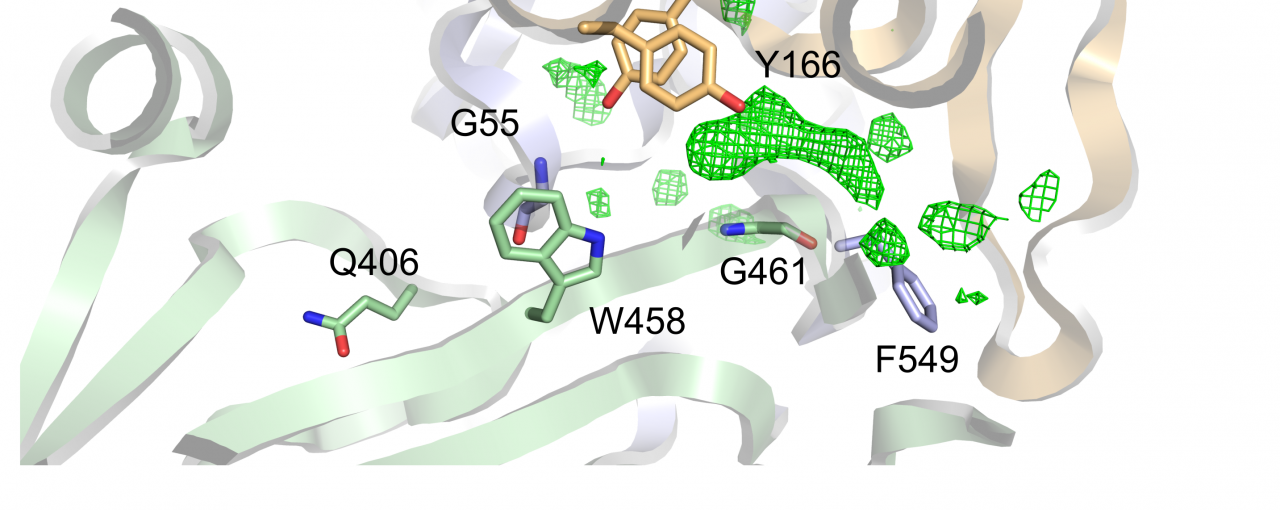
Scientists at Research Complex, Diamond Light Source and University of Oxford have focused on determining the structure of SapA - a protein that was proposed to deliver heme and AMPs to the transmembrane Sap component. With the structure of SapA determined they could directly assess the proteins ability to interact with heme and AMPs. While they found evidence for weak heme binding at the molecular surface of SapA, they also found that the buried ligand binding cavity of SapA was of insufficient volume to accommodate heme or folded AMPs. Instead, based on other analysis and experiments, they proposed that the cavity is most likely occupied by a hydrophobic di- or tri- peptide. Unexpectedly, they also discovered that a double stranded ribonucleic acid (RNA) molecule coordinates with the SapA protein both in the crystal structure and in solution. Whilst the role of this interaction remains unclear, the scientists found that the binding affinity of the RNA to SapA is relatively strong and within the range observed for other known RNA/protein complexes and could therefore be physiologically significant.
Overall, the authors explain in molecular detail the interactions of SapA with heme and dsRNA and propose a role for SapA in the transport of di- or tri-peptides that provides a source of nutrients and is linked to virulence. Further work is needed to fully understand the role of the NTHi Sap transporter but this work lays the groundwork to direct efforts to investigating other mechanisms and pathways that could explain the previously observed heme requirement and AMP sensitivity of NTHi Sap transporter.
Martin Walsh, Group Leader at Research Complex and Deputy Director of Life Sciences at Diamond Light Source says,
The role of the SAP transporter and indeed peptide transporters in the virulence of gram negative bacteria is still poorly understood. Structural data can provide key insights and validate hypotheses that have been derived by modelling or other indirect methods. Our work on SapA is a good starting point for understanding how it contributes to NTHi evading our innate immune response.
Petra Lukacik, Project Lead and Research Scientist at Diamond Light Source adds:
With this work we set out with a strong vision on what the results should be. Luckily science does not work like that. Not only has our structural study illuminated data obtained by other biological groups, it has also yielded some unexpected surprises. With the structure of SapA we have added a piece to the jigsaw puzzle that is the biology of a clinically important pathogen.
Lukacik, P., Owens, D.L., Harris, G., Reddy Bolla, J., Picaud, S., Alibay, I., Nettleship. J.E., Bird, L.B., Owens, R.J., Biggin, P.C., Filippakopoulos, P., Robinson, C.V. and Walsh, M., The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif, PLOS One (2021). DOI: https://doi.org/10.1371/journal.pone.0256070
Image: Fig 3. The SapA ligand-binding cavity. A, a detailed view of the SapAclosed ligand binding cavity. Aromatic residues lining the cavity are shown in stick representation together with the other residues discussed in the text. Unexplained Fo-Fc residual density is represented as a green mesh and is contoured at 3σ. B, left: The same Fo-Fc density highlighted in the context of the overall view of SapAclosed (cα trace). Right: The ligand-binding cavity of SapAclosed as calculated by the program Voidoo (magenta mesh). Arrows indicate cavity openings to the protein surface.