Despite the success of vaccination programs throughout the world, COVID-19 remains a threat to human health at the beginning of 2022. The development of safe and efficient small-molecule therapeutics to prevent or treat acute viral infections is of tremendous importance, as antiviral drugs are often designed to suppress the activity of human or viral proteins, including proteases.
Proteases are an important class of enzymes, which catalyse the hydrolysis of proteins and peptides into smaller fragments. Several viruses encode proteases in their genome, which often have essential functions in the viral replication cycle. The SARS-CoV-2 genome, for example, encodes for two proteases, the main protease (Mpro) and the papain-like protease (PLpro), which are essential for viral replication as they process the initially translated SARS-CoV-2 polyprotein into functional proteins. Drug discovery programs have mainly focused on developing small-molecule Mpro inhibitors to suppress viral replication, resulting in the recent approval of nirmatrelvir (co-administered with ritonavir and marketed as paxlovid) for COVID-19 treatment.
In addition to processing the viral polypeptide chain, PLpro also catalyses the fragmentation of human proteins involved in the innate immune response, a function which helps SARS-CoV-2 in evading this very response. Due to this dual mode of action of PLpro, the development of small-molecule antivirals that efficiently suppress PLpro activity appears to be particularly attractive to treat COVID-19. However, the development of such small-molecules is difficult, partly because of a lack in accurate and simple methods to measure the activity of PLpro in the laboratory, which is essential to assess the effect of small-molecules on PLpro.
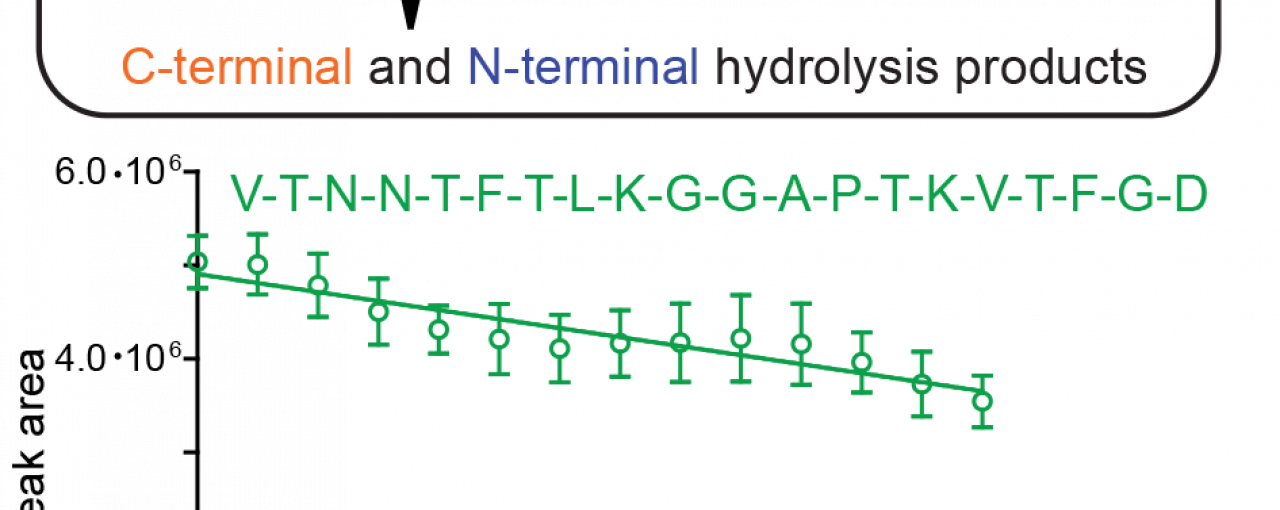
Building on a collaboration established to advance SARS-CoV-2 research, scientists at the University of Oxford, Diamond Light Source, and the Research Complex at Harwell have developed an efficient mass spectrometry-based method to measure the protease activity of SARS-CoV-2 PLpro in the laboratory. A simple oligopeptide, that mimics a natural PLpro substrate, is exposed to isolated PLpro which catalyses the cleavage of the oligopeptide into two fragments of reduced masses. The formation of the two lighter fragments is directly measured as a function of time using mass spectrometry and compared to an internal standard, which enables the quantification of fragment formation and thus PLpro activity.
The PLpro -catalysed oligopeptide fragmentation can be performed in the presence of small-molecules; this experimental set-up can quantify the extent to which small-molecules suppress PLpro activity. The mass spectrometry-based method thus complements established fluorescence-based methods, which are widely-used to investigate the effect of small-molecules on PLpro activity. Importantly, the research revealed that several, but not all, small-molecules, which have been reported to suppress PLpro activity when using fluorescence-based methods, did not suppress PLpro activity when employing the novel mass-spectrometry-based method. This might be a consequence of the more biologically-representative oligopeptide used, which likely binds PLpro more efficiently than the substrates commonly used in fluorescence-based methods; small-molecules competing with the substrate for binding PLpro will do so less efficiently for a better substrate.
This research highlights the importance of using complementary experimental methods to validate the effect of small-molecules on PLpro activity. It can be expected, that the novel mass spectrometry-based method will accelerate drug discovery campaigns aimed at identifying small-molecules which efficiently suppress SARS-CoV-2 PLpro activity.
Lennart Brewitz, Jos J. A. G. Kamps, Petra Lukacik, Claire Strain Damerell, Yilin Zhao, Anthony Tumber, Tika R. Malla, Allen M. Orville, Martin A. Walsh and Christopher Schofield, Mass spectrometric assays reveal discrepancies in inhibition profiles for the SARS‐CoV‐2 papain‐like protease, ChemMedChem (2022).
Figure from article:
A mass spectrometry-based direct SARS-CoV-2 PLproassay reveals discrepancies in inhibition profiles for non-covalent inhibitors. Use of assays orthogonal to the established fluorescence-based assays is important for development of efficient small-molecule PLproinhibitors.