The damage caused by Covid-19 to the lungs’ smallest blood vessels has been intricately captured using high-energy X-rays emitted by a special type of particle accelerator.
The Research Complex at Harwell and scientists from University College London (UCL) and the European Synchrotron Research Facility (ESRF) have used a new revolutionary imaging technology called Hierarchical Phase-Contrast Tomography (HiP-CT), to scan donated human organs, including lungs from a Covid-19 donor.
HiP-CT enables 3D mapping across a range of scales, allowing clinicians to view the whole organ as never before by imaging it as a whole and then zooming down to cellular level.
The technique uses X-rays supplied by the European Synchrotron (a particle accelerator) in Grenoble, France, which following its recent Extremely Brilliant Source upgrade (ESRF-EBS), now provides the brightest source of X-rays in the world at 100 billion times brighter than a hospital X-ray.
Due to this intense brilliance, researchers can view blood vessels five microns in diameter (a tenth of the diameter of a hair) in an intact human lung. A clinical CT scan only resolves blood vessels that are about 100 times larger, around 1mm in diameter.
Dr. Claire Walsh (UCL Mechanical Engineering) said:
The ability to see organs across scales like this will really be revolutionary for medical imaging. As we start to link our HiP-CT images to clinical images through AI techniques, we will - for the first time - be able to highly accurately validate ambiguous findings in clinical images. For understanding human anatomy this is also a very exciting technique, being able to see tiny organ structures in 3D in their correct spatial context is key to understanding how our bodies are structured and how they therefore function.
Using HiP-CT, the research team, which includes clinicians in Germany and France, have seen how severe Covid-19 infection ‘shunts’ blood between the two separate systems – the capillaries which oxygenate the blood and those which feed the lung tissue itself. Such cross-linking stops the patient’s blood from being properly oxygenated, which was previously hypothesised but not proven.
Dr. Paul Tafforeau, lead scientist at ESRF, said:
The idea to develop this new HiP-CT technique came after the beginning of the global pandemic, by combining several techniques that were used at the ESRF to image large fossils, and using the increased sensitivity of the new Extremely Brilliant Source at the ESRF, ESRF-EBS. This allows us to see in 3D the incredibly small vessels within a complete human organ, enabling us to distinguish in 3D a blood vessel from the surrounding tissue, and even to observe some specific cells.
This is a real breakthrough, as human organs have low contrast and so are very difficult to image in detail with the current available techniques. ESRF-EBS has allowed us to go from deciphering the secrets of fossils to seeing the human body as never before.
Using HiP-CT to create the Human Organ Atlas
With support from the Chan Zuckerberg Initiative (CZI), the UCL-led team are using HiP-CT to produce a Human Organ Atlas, launching this week. This will display six donated control organs: brain, lung, heart, two kidneys and a spleen, and the lung of a patient who died of Covid-19. There will also be a control lung biopsy and a Covid-19 lung biopsy. The Atlas will be available online for surgeons, clinicians and the interested public.
Project lead Professor Peter Lee (UCL Mechanical Engineering) and Leader of the Materials Design & Manufacturing Group at Research Complex said:
The Atlas spans a previously poorly explored scale in our understanding of human anatomy, which is the centimetre to micron scale in intact organs. Clinical CT and MRI scans can resolve down to just below a millimetre, whilst histology (studying cells / biopsy slices under a microscope), electron microscopy (which uses an electron beam to generate images) and other similar techniques resolve structures with sub-micron accuracy, but only on small biopsies of tissue from an organ. HiP-CT bridges these scales in 3D, imaging whole organs to provide new insights into our biological makeup.
Insights for other diseases and conditions
The researchers are confident that the scale-bridging imaging from whole organ down to cellular level could provide additional insights into many diseases such as cancer or Alzheimer’s Disease.
It is hoped that the Human Organ Atlas will eventually contain a library of diseases that affect organs on a range of scales, from 1 to 100s of microns to entire organs, helping clinicians as they diagnose and treat a wide range of diseases.
The team also hope to use machine learning and artificial intelligence to calibrate clinical CT and MRI scans, enhancing the understanding of clinical imaging and enabling faster and more accurate diagnosis.
The work, published in Nature this week, was supported by the Chan Zuckerberg Initiative, the ESRF, the UK-MRC, the Research Complex at Harwell and the Royal Academy of Engineering. Additional support was from the German Centre for Lung Research (DZL, BREATH), the ERC, the German Registry of COVID-19 autopsies (DeRegCOVID), INSERM, University of Grenoble Alpes, Kidney Research UK, Rosetrees Trust, the Wellcome Trust, GOSH and the German Registry of COVID-19 Autopsies.
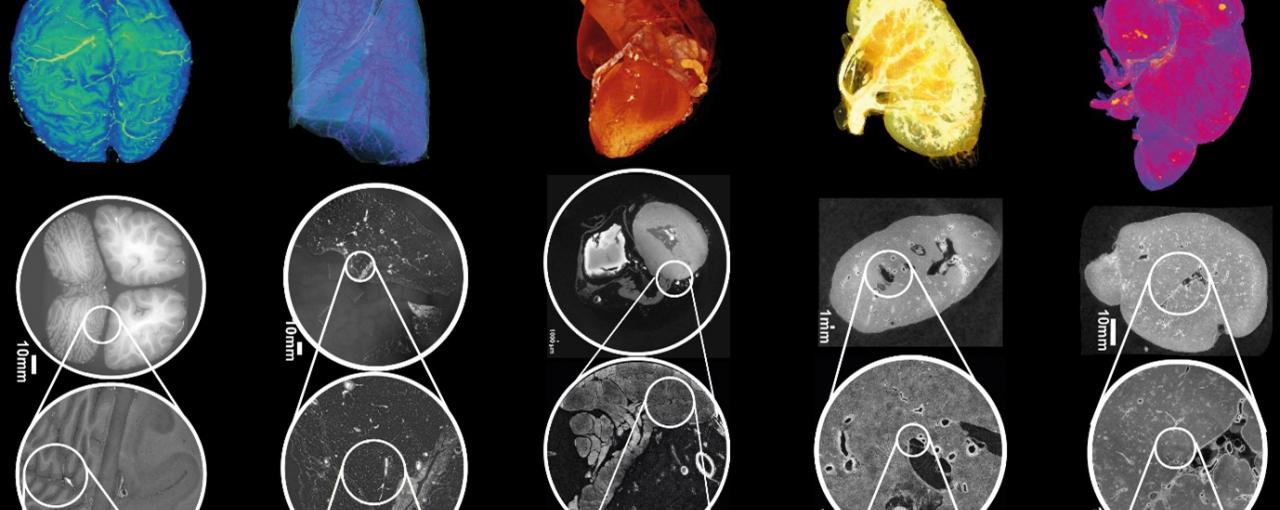
Image: Figure 2: HiP-CT of brain (a), lung (b), heart (c) kidney (d) and spleen (e); for each organ, 3D rendering (i) of the whole organ is shown using scans at 25 µm per voxel. Subsequent 2D slices (ii–iv) show positions of the higher-resolution VOI relative to the previous scan. (v), Digital magnification of the highest-resolution image with annotations depicting characteristic structural features in the brain (ml, molecular layer; gl, granule cell layer; pc, Purkinje cell), in the lung (c, blood capillary; ec/m, epithelial cell or macrophage), in the heart (mc, myocardium; ca, coronary artery; ad, adipose tissue), in the kidney (e/a, efferent or afferent arteriole; g, glomerulus) and in the spleen (rp, red pulp; wp, white pulp; a, arteriole; ss, splenic sinus). All images are shown using 2× binning.